
Getting Started with cogxwalkr
getting-started.rmdIntroduction
cogxwalkr is an R package designed to facilitate
crosswalking—or translating effect estimates—between different cognitive
outcome measures in studies of cognitive aging, exposures, and
interventions. It enables researchers to compare results across studies
that use different cognitive tests.
This package implements methods described in (Ackley et al. 2025). The manuscript authors developed two methods to crosswalk estimated treatment effects of cognitive outcomes that are flexible, broadly applicable, work when only summary statistics are available, and do not rely on strong distributional assumptions. This method requires access to a dataset that includes both the cognitive measure you wish to crosswalk from and the cognitive measure you wish to crosswalk to.
The package was developed by Sarah Ackley and Jason Gantenberg. This vignette demonstrates how to use cogxwalkr in R.
Installing
To install the package, you will first need to install and load the remotes package, which you can do from the R console:
To install the package:
install.packages("remotes")
remotes::install_github("jrgant/cogxwalkr")Installation Errors
You may get an error message that reads:
Using GitHub PAT from the git credential store.
Error: Failed to install 'remotes' from GitHub:
HTTP error 401.
Bad credentialsThe most likely cause is that remotes detects a bad
Github token in your credential store. Run
gitcreds::gitcreds_delete(), select option 2
to remove the bad credential from the store, and rerun the
remotes::install_github() command above.
Alternatively, you may get a similar error message that reads:
Using github PAT from envvar GITHUB_PAT. Use `gitcreds::gitcreds_set()` and unset GITHUB_PAT in .Renviron (or elsewhere) if you want to use the more secure git credential store instead.
Error: Failed to install 'cogxwalkr' from GitHub:
HTTP error 401.
Bad credentialsIn this case, run Sys.setenv(GITHUB_PAT = "") and then
rerun the remotes::install_github() command above.
Motivation
As a motivating example, consider a study estimating the effect of
APOE-ε4 carrier status on scores from the MMSE (Mini-Mental State
Examination). If we wish to compare this effect to results from another
study that used the MoCA (Montreal Cognitive Assessment), we need a
conversion factor that allows us to convert a summary measure on the
MMSE scale to the MoCA scale. cogxwalkr aids in estimating
the conversion coefficient required to crosswalk from the MMSE to the
MoCA under the following general assumptions regarding two cognitive
measures (Ackley et al. 2025):
- they measure the same underlying construct (e.g., executive function, global cognition, etc.)
- they are correlated only through their measurement of this common construct.
Usage
To load the package:
Adjunct Data
These crosswalk methods require individual-level data from an adjunct
study: a study where both cognitive measures are performed for each
participant. In this package, the built-in cogsim dataset
represents the adjunct study data. The adjunct data is simulated data
based on the Alzheimer’s Disease
Neuroimaging Initiative (ADNI). This dataset includes an MMSE score
and MoCA score conditional on binary dementia status (0 for absent and 1
for present) for approximately 1500 study participants. Code used to fit
and simulate these data can be found at .
To view the example adjunct study, cogsim, we can type
either of the following into the R console:
Additional information is available via ?cogsim.
We implement two methods to obtain crosswalk slopes: 1) unconditional method and 2) conditional method. The unconditional method requires only data on both cognitive measures for each individual, whereas the conditional method requires additional data on a third binary variable, here dementia status.
Unconditional Method
To obtain an estimate of the crosswalk slope using the unconditional method we can use the crosswalk function, which is the workhorse of the package:
crosswalk(cog1 = "mmse", cog2 = "moca", data = cogsim, niter = 500)This estimates a crosswalk slope from cog1, here the
MMSE, to cog2, here the MoCA, using the built-in cogsim
data as the adjunct data. cog1 and cog2 must
be supplied as strings. niter gives the number of random
splits used to obtain the crosswalk. The built-in cogsim
dataset can be replaced with any other dataset containing the
appropriate columns. Since this uses the adjunct data without any
bootstrap resampling, no uncertainty on the crosswalked slope is
presented.
crosswalk(cog1 = "mmse",
cog2 = "moca",
data = cogsim,
niter = 500,
control = list(nboot = 100, seed = 123))
crosswalk(cog1 = "mmse",
cog2 = "moca",
data = cogsim,
niter = 500,
control = list(nboot = 100, seed = 123456, ncores = 8))Unconditional Method Using Summary Statistics
The unconditional method can also be used based only on summary statistics. (In many cases, it may be more statistically efficient to do so, as the number of unconditional split iterations must be chosen with care.)
The summary statistics approach is accessible directly via the
est_cw_coef() function.
# estimate slope using lm()
est_cw_coef(cog1 = "mmse", cog2 = "moca", data = cogsim, method = "lm")
# estimate slope using manual calculation
est_cw_coef(cog1 == "mmse", cog2 = "moca", data = cogsim, method = "manual")However, we recommend using crosswalk() because other
functions in cogxwalkr are built to act upon its output,
not that of est_cw_coef().
Omitting the niter argument when calling
crosswalk() will result in the package using the “lm”
method to estimate the crosswalk:
crosswalk(cog1 = "mmse", cog2 = "moca", data = cogsim)Conditional Method
To use the conditional method, we must specify an additional binary
variable on which to perform the conditional splits. The optional
condition_by input declares this input and, when present,
will elicit the conditional method. Note that crosswalk()
will ignore niter if condition_by is
specified.
crosswalk(cog1 = "mmse",
cog2 = "moca",
data = cogsim,
condition_by = "dementia")Summary and Plotting Functions
Summaries
crosswalk() outputs a list containing the results of the
estimation procedure,1 along with information about the
specifications selected by the user. Similar to glm() and
lm(), the cogxwalkr package comes with
functions to help summarize results.2
boot_settings <- list(nboot = 100, seed = 999, ncores = 1)
# summary statistics estimation
cws <- crosswalk(cog1 = "mmse",
cog2 = "moca",
data = cogsim,
control = boot_settings)
#> `ncores` is set to 1 (default). To use parallel processing, set `ncores` to the desired number of cores or to 999 to use the maximum available.
#> Running bootstraps over 1 cores ...
summary(cws)
#>
#> --------------------------------------------------
#> Crosswalk Summary (Adjunct)
#> --------------------------------------------------
#> Formula: moca ~ mmse
#> Coefficient: 1.247
#>
#> 95% confidence limits:
#> (1.170, 1.323) - normal
#> (1.174, 1.317) - percentile
#>
#> Based on 100 bootstrap replicates
#> SE = 0.0391
#> --------------------------------------------------
#> Number of iterations:
#> Conditioning variable:
# unconditional split
cwu <- crosswalk(cog1 = "mmse",
cog2 = "moca",
data = cogsim,
niter = 500,
control = boot_settings)
#> `ncores` is set to 1 (default). To use parallel processing, set `ncores` to the desired number of cores or to 999 to use the maximum available.
#> Running bootstraps over 1 cores ...
summary(cwu)
#>
#> --------------------------------------------------
#> Crosswalk Summary (Adjunct)
#> --------------------------------------------------
#> Formula: moca ~ mmse - 1
#> Coefficient: 1.154
#>
#> 95% confidence limits:
#> (1.012, 1.296) - normal
#> (1.105, 1.375) - percentile
#>
#> Based on 100 bootstrap replicates
#> SE = 0.0725
#> --------------------------------------------------
#> Number of iterations: 500
#> Conditioning variable:
# conditional split
cwc <- crosswalk(cog1 = "mmse",
cog2 = "moca",
data = cogsim,
condition_by = "dementia",
control = boot_settings)
#> `ncores` is set to 1 (default). To use parallel processing, set `ncores` to the desired number of cores or to 999 to use the maximum available.
#> Running bootstraps over 1 cores ...
summary(cwc)
#>
#> --------------------------------------------------
#> Crosswalk Summary (Adjunct)
#> --------------------------------------------------
#> Formula: moca ~ mmse - 1
#> Coefficient: 1.389
#>
#> 95% confidence limits:
#> (1.290, 1.489) - normal
#> (1.299, 1.496) - percentile
#>
#> Based on 100 bootstrap replicates
#> SE = 0.0506
#> --------------------------------------------------
#> Number of iterations: 204
#> Conditioning variable: dementiaThe outputs of summary() can be stored as objects
(lists). To view the raw form of such an object, use
print.AsIs().
See the documentation for summary.cogxwalkr() for
information on options concerning the
level and method used to estimate bootstrapped confidence intervals.
Plotting
In general, we leave plotting the output, results, etc., up to the user. The package does provide simple plot output to allow users to review interim results quickly.
We will use the cwu object we saved in the prior section
as an example. This object contains the results of a crosswalk
estimation that used the unconditional split method, along with
bootstrapping to estimate 95% confidence intervals. The
cxsum argument takes a summary output and overlays these
confidence intervals.
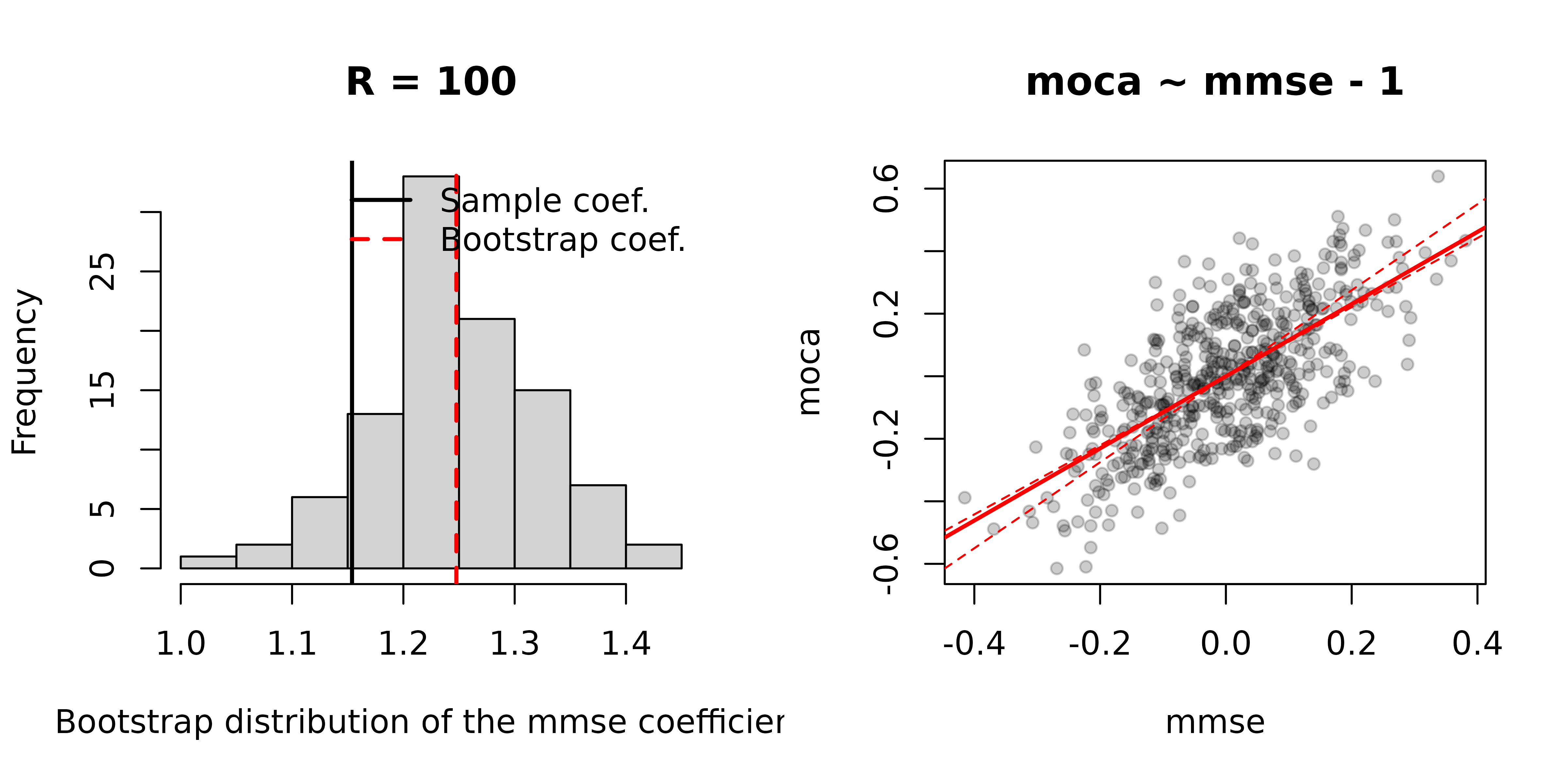
Alternatively, if we used the summary statistics method
(cws above), we will see something a bit different. We no
longer have a plot of the split differences.
plot(cws, lcex = 0.7)
#> The crosswalk() object does not contain differences, most likely because the slope was calculated using the manual method and not unconditional splits. The scatterplot has been omitted.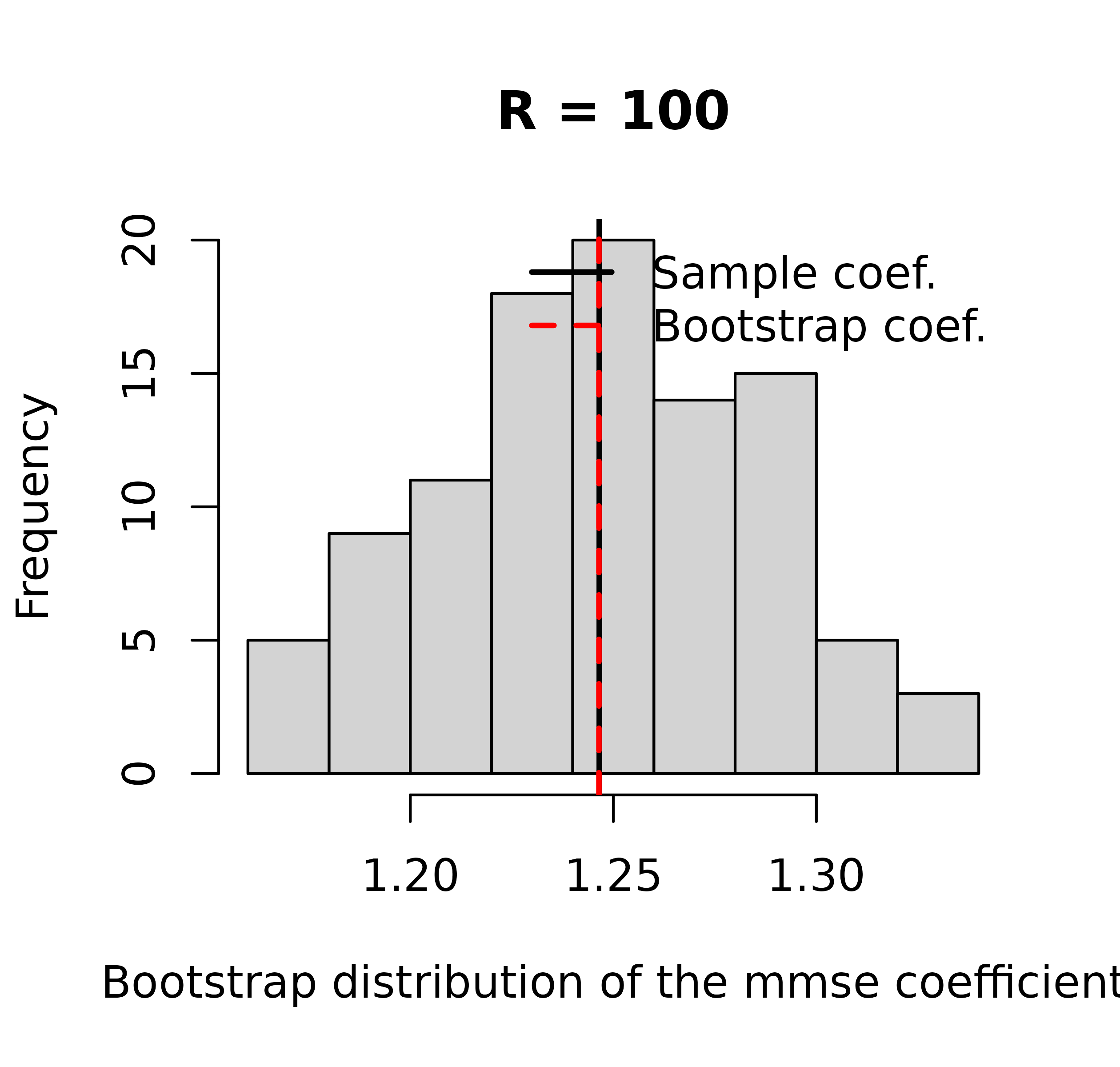
Finally, if we conducted a splitting method without bootstrapping, we can suppress the bootstrap plot panel as follows.
cwu_noboot <- crosswalk(cog1 = "mmse", cog2 = "moca", data = cogsim, niter = 500)
plot(cwu_noboot, types = "slope")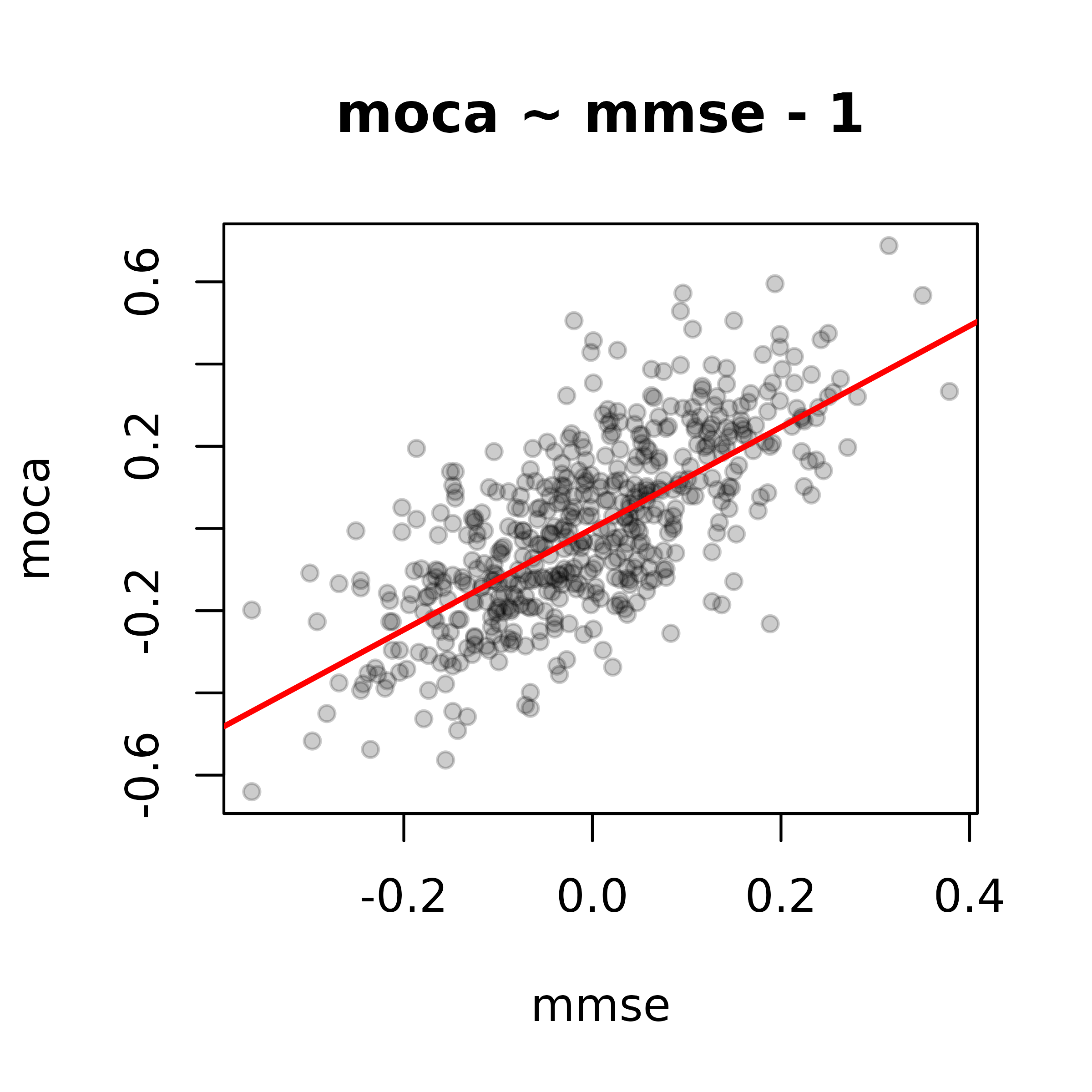
As with, summary.cogxwalkr(), additional options for
plotting are available in the documentation for
plot.cogxwalkr().
Crosswalking
Let’s say you have estimated a crosswalk from MMSE to MoCA in your
adjunct data. We’ll use cwu for this example. You come
across summary data from another (fictional) study that describes the
difference in MMSE for those with APOE-ε4 carrier status versus not
(-2.4, SE = 0.064, 95% CL: [-2.52, -2.27]) and would like to know what
the difference on the MoCA would be for this sample.
In that case, you can use do_crosswalk() in one of two
ways to obtain this estimate: using either the standard error from the
summary data or the confidence interval.
pub_outcome_label <- "mmse"
pub_pred_label <- "apoe4"
# using the standard error
dcw_se <- do_crosswalk(cwu, est_mean = -2.4, est_se = 0.064,
est_indep = pub_pred_label, est_outcome = pub_outcome_label)
# using the confidence interval
dcw_ci <- do_crosswalk(cwu, est_mean = -2.4, est_ci = c(-2.52, -2.27),
est_indep = pub_pred_label, est_outcome = pub_outcome_label)
# results will differ slightly due to rounding error
dcw_se
#>
#> --------------------------------------------------
#> Crosswalk Summary
#> --------------------------------------------------
#> Adjunct Estimate:
#> 1.154, SE: 0.0537
#> moca ~ mmse - 1
#>
#> Study Estimate:
#> -2.400, SE: 0.064
#> mmse ~ apoe4
#>
#> Crosswalked Estimate:
#> -2.769, SE: 0.149
#> 95% confidence limits: (-3.060, -2.478)
#> --------------------------------------------------
dcw_ci
#>
#> --------------------------------------------------
#> Crosswalk Summary
#> --------------------------------------------------
#> Adjunct Estimate:
#> 1.154, SE: 0.0537
#> moca ~ mmse - 1
#>
#> Study Estimate:
#> -2.400, SE: 0.0638
#> mmse ~ apoe4
#>
#> Crosswalked Estimate:
#> -2.769, SE: 0.148
#> 95% confidence limits: (-3.060, -2.478)
#> --------------------------------------------------The crosswalk element in the list returned contains an
estimated mean, standard error, and
()%
confidence interval (by default
),
accounting for uncertainty in both the crosswalk estimate and published
summary.3
In this case, we see the estimated difference in MoCA comparing APOE-ε4
carriers to non-carriers is -2.77 (SE = 0.149, 95% CI: [-3.06, -2.48]),
based on the information in dcw_se.